Microbiology
Contents
Here we describe how bacterial, fungal and viral infections are identified, using a variety of samples and biochemical and biological laboratory techniques. We will list some of the infectious agents most relevant to oral and maxillofacial surgery in each category. The corresponding infections and conditions as well as their treatment by surgery and/or medication are described elsewhere.
Specimens suitable to identify infectious agents can be blood (most common) or urine samples, or any other body fluid, including pus, as well as skin or hair samples. Samples may be obtained from swabs of various body surfaces, or may be obtained from a biopsy. The specimen needed varies for different suspected diagnoses. Identifying (and quantifying) the cause of an infection is important for successful treatment planning and execution: speed of action can be essential and so is the prompt choice of the most suitable antibiotic regimen for bacterial infections. Severe infections, with bacterial resistance to many mainstream antibiotics on the rise, can and do kill. Having said that, the evidence to date supports “best guess” antibiotic treatment in head and neck infections.
Viral infections
Searching and / or screening for viral infections requires a different approach from testing for fungal and most bacterial infections. A virus is not able to replicate without using the host’s cellular replication mechanisms. That means that it is difficult to grow a culture of a sample of a suspected virus. Instead, the analysis has to rely on the host’s immune system response to the infection, or more recently on techniques that allow to multiply small virus DNA sections in a sample.
Antibodies
When fighting infections, the body’s immune system produces immunoglobulins (antibodies). There are different families of antibodies, found in different parts of the body, with a range of tasks in the immune system and in variable quantities. Enhanced levels of such general types of antibodies will strongly indicate an infection (past and/or present). Quantifying the levels of the main antibody types will usually just be the first step in identifying a particular viral infection. Each viral infection will lead to the production of highly specific antibodies. Thus identifying an infection by a particular virus requires looking for antibodies specific to that virus. Again, past and successfully overcome infections will leave a specific antibody fingerprint in the host’s blood (which is re-activated upon renewed infection which will be promptly erased, the basis of immunity) and so do successful immunizations. In fact, monitoring the success of vaccination / immunization schemes relies on finding specific, pre-determined levels of antibodies, the production of which was triggered by the immunization. It is normally not difficult to identify if somebody suffers from an acute viral infection but some such infections are more latent and low-key, not showing signs of an acute illness.
PCR
Being able to identify and quantify the presence of a particular virus in a patient is desirable, rather than having to rely on the evidence from antibodies. Such direct identification techniques do exist and have become the gold standard in identifying viral (and other) infections. The technique employs the polymerase chain reaction (PCR) which was originally developed in genomics research but quickly became an invaluable diagnostic tool.
For PCR to work the DNA sequence of the infectious agent has to be known. A small fragment of the known sequence is added to the sample, alongside enzymes, nucleotides and other components for the reaction. If the sample contains DNA with an identical section to that of the small kick-start amount added, then that DNA will be multiplied by a very large factor (up to about 106 times) quickly. In the absence of such a match, no multiplication occurs. Having to know the DNA sequence of an infectious agent and having to have a small quantity of a primer sample to get the reaction started in 2017 is no longer a hurdle.
PCR has a number of advantages: it is quick, it is able to return quantitative results (‘viral load’), it does not require special equipment (or skills), it is relatively low cost and is well suited to be completely automated in the not too distant future. In fact, there are predictions that PCR will soon become a standard part of a routine dental check-up with regard to periodontal disease and dental decay for identification of a range of bacterial infections.
Some viral infections that are relevant in a maxillofacial context:
- Coxsackie virus (RNA virus; thought to be involved in the development of Sjögren’s syndrome).
- Cytomegalovirus (very common; belongs to the family of herpes viruses, often no symptoms of infection but is a cause for concern in immune-compromised people).
- Epstein-Barr virus (acutely causing glandular fever; suspected to be the cause of hairy leukoplakia via opportunistic infections in immune-compromised patients (after transplantations, or HIV infections)).
- Hepatitis B, C (there are five main types of hepatitis. Type A is usually self-limiting but highly contagious and is transmitted mainly by contaminated food and water. Types B and C are transmitted by blood and other body fluids. Both can be self-limiting but are more likely to lead to lifelong infections that can lead to liver disease and liver malignancy; effective antiviral medications to treat different forms of hepatitis are available, hepatitis C treatment has become available very recently).
- Herpes simplex viruses 1 (cold sores) and 2 (genital herpes) are ubiquitous and contagious. Both invade, and lie dormant in, nerves, particularly the trigeminal nerve.
- HIV, human immunodeficiency virus (a RNA retrovirus; untreated infection long-term leads to immunodeficiency syndrome, AIDS).
- HPV (16 & 18), human papilloma virus (about 200 types known, affect mostly skin (warts) and mucosa. Some are high-risk infections for causing a variety of cancers, in particular types 16 and 18 are responsible for some type of oral and oropharyngeal cancers, whereas types 6 and 11 are responsible for the growth of some benign tumours in the respiratory tract.
- Kaposi’s sarcoma virus (another herpes virus, closely related to Epstein-Barr virus; causes lymphoma and Kaposi’s sarcoma (a particular type of cancer of blood vessels) in people with AIDS)
- Paramyxovirus (mumps; a RNA virus)
- Varicella (chickenpox; one of several herpes viruses that infect humans; after the initial infection the virus goes dormant in nerve tissue, including cranial nerves. Later in life it may reactivate in the form of herpes zoster (shingles)).
Parasitic infections
Infections with parasites are not likely to be a major topic in a maxillofacial clinic, the only exception perhaps being toxoplasma gondii, a single-cell parasite that is for part of its life cycle hosted by cats and causes symptoms known as toxoplasmosis in its human hosts in other periods of its life cycle.
Fungal infections
Identification of fungal infectious species rarely requires any further sample preparation, most species can be directly identified under a microscope: spores and fungi have characteristic shapes. If not, then a culture of the specimen can be grown. A culture is grown by infecting a suitable sterile nourishing medium with the sample taken, and then keep the infected medium under optimal growing conditions (temperature, humidity). If a fungal colony grows, the colony will grow in shapes and patterns that are characteristic for the fungus type (there may be more than one present). Many fungal colonies can be identified by their macroscopic colony patterns, or else by microscopy.
Some fungal infections that are relevant in a maxillofacial context:
- Aspergillus: a very common mould; of the many aspergillus species only few infect humans and if so, it is usually only problematic for immune-compromised people.
- Candida albicans: a yeast infecting humans, causing candidiasis (thrush). Infection can be systemic and serious in immune-compromised people (HIV-infected, after transplants or chemotherapy) and in the form of hospital-acquired infections. C. albicans biofilms can infect the surface of implanted medical devices. The most common location of candidiasis is the oral mucosa, with lack of oral hygiene, prolonged and high-dose antibiotic treatments and dry mouth increasing the risk for C. albicans infections.
- Candida dublensii: a less common form which is resistant to the regular treatment of fluconazole.
Bacterial infections
Bacterial infections constitute a very wide field and there are legions of different bacteria, some pathogenic, some commensal (in peaceful coexistence with the host).
There are several ways in which bacteria are classified. One way is by their behaviour when staining a specimen with a particular dye, crystal violet. This dye distinguishes different kinds of cell walls in different bacteria. This is illustrated in Figure 1.
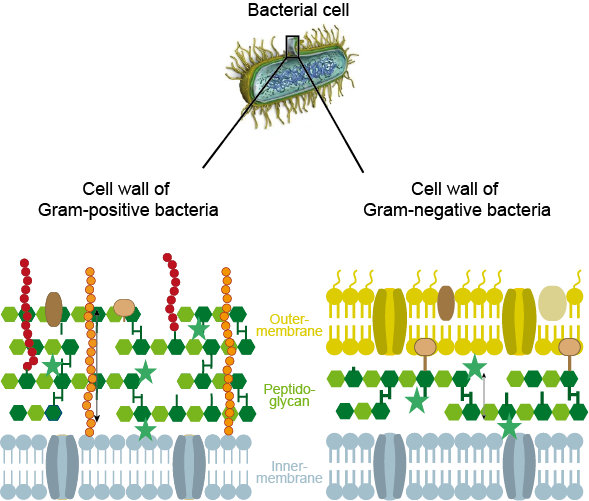
Gram-positive bacteria retain the crystal violet dye after washing because the dye is firmly bonded to the peptidoglycan outer layer of these bacterial cell walls. Gram-negative bacteria have a second outer cell wall membrane which does not strongly interact with crystal violet dye, so will not be stained because the dye is easily washed off (there are also some types of bacteria that do not neatly fit one of the two categories). This categorisation is named after the Danish scientist H.C. Gram who developed this staining technique. The different types of cell walls also have an impact on treatment options for different types of bacteria.
Different bacteria have different shapes, so bacterial morphology (under a microscope) is helpful in identification as well. By and large, cocci (round shaped) are most common amongst Gram-positive bacteria, Gram-negative bacteria are often rod shaped.
Another way to classify bacteria is by the environment in which they grow, aerobic or anaerobic. Some bacteria can only live in anaerobic conditions and oxygen is toxic for them (some of our helpful gut bacteria belong to that category), other types of bacteria need oxygen for their metabolism and thus can only live in aerobic conditions (there are also some types of bacteria that fall in between these two categories; for example facultative anaerobic bacteria can tolerate some levels of oxygen).
Tables 1 – 3 give an overview of these different types of bacteria: each of the named categories has many more subspecies.
Table 1 Gram-positive bacteria
| Name of species | Shape | Oxygen needs |
|---|---|---|
| Actinomyces | irregular | anaerobic |
| Bacilli | rods | aerobic |
| Clostridia | rods, spores | anaerobic |
| Enterococci | cocci | facultative anaerobic |
| Listeria | rods | facultative anaerobic |
| Pneumococci | irregular | facultative anaerobic |
| Staphylococci | cocci | facultative anaerobic |
| Streptococci | cocci | facultative anaerobic |
Table 2 Gram-negative bacteria
| Name of species | Shape | Oxygen needs |
|---|---|---|
| bacteroides | rods | anaerobic |
| bartonella | rods | aerobic |
| campylobacter | rods | aerobic |
| enterobacteriaceae (e.coli; klebsiella, shigella, salmonella) | rods | facultative anaerobic |
| haemophilus | irregular | facultative anaerobic |
| legionella | rods | aerobic |
| neisseria | cocci | aerobic |
| pseudomonas | rods | aerobic |
Table 3 Miscellaneous bacteria
| Name of species | Shape | Oxygen needs |
|---|---|---|
| borrelia | irregular | -- |
| heliobacter | irregular | aerobic |
| mycobacteria | rods | aerobic |
| mycoplasma | irregular (no cell wall) | -- |
| rickettsia | irregular | -- |
The work-up in the identification process of many bacterial infectious agents is similar to the routines used in fungal infections: infect a sterile growing medium with the specimen, and keep in optimal growing conditions. Once a colony of bacteria has grown, staining and microscopy are standard ways of identification. That is all well-established and works for many types of bacteria. It does not work for bacteria that are hard or impossible to cultivate (many anaerobic bacteria) or which grow very slowly (for example, the bacteria causing tuberculosis). For these previously difficult to identify species, nowadays PCR methods (see above) are available, similar to the techniques used to identify viruses based on their DNA (or RNA).
Being able to grow cultures of bacterial colonies is not only important to identify the infectious agent(s), it is also necessary to find out about sensitivity to various antibiotics. With increasing problems caused by bacterial strains that are resistant to main-stream antibiotics (for example, MRSA (methicillin-resistant staphylococcus aureus) infections), this aspect becomes increasingly important.