Infection
Contents
Below we describe in slightly more technical detail some commonly used antibiotic drugs to treat bacterial, fungal and viral infections. Our discussion will also include the timely topic of resistance to antibiotic drugs.
Antibacterial agents
With a wide range of bacterial infections being common problems encountered in a maxillofacial clinic, the prescription and use of antibacterial agents are common practice. Despite this, the number of different antibacterial agents routinely used in oral and maxillofacial surgery is fairly small but nevertheless includes examples of mechanisms from all four categories of antibacterial interferences, interfering with the bacterial cell wall, bacterial metabolism, bacterial protein synthesis or bacterial DNA.
Antibacterial agents that interfere with the bacterial cell wall
The best known representatives of this class of agents are probably the so called β-lactams, penicillin and its many derivatives. The general structure is shown in Figure 1 where the basic functional unit, the β-lactam 4-membered ring structure, is highlighted in red.
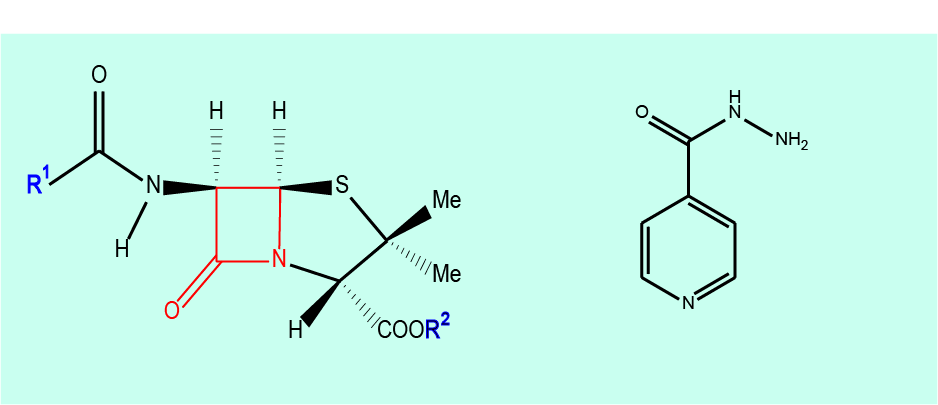
Widely used drugs from this group are penicillin (V & G), ampicillin, amoxicillin, flucloxacillin, methicillin, several cephalosporin derivatives and clavulanic acid. The β-lactam derivative clavulanic acid itself is not an antibacterial agent but in combination with other β-lactam drugs it can overcome resistance (see below) in bacteria which would otherwise be able to inactivate β-lactam drugs by producing an enzyme, β-lactamase. Co-amoxiclav is the best known example.
Isoniazid (see right part of Figure 1) is a different type of chemical but also belongs to the class of antibacterial agents that alter the stability of the bacterial cell wall. It has no clinical role in maxillofacial surgery.
A less widely used, last-resort antibacterial agent is vancomycin (a natural product, isolated from some soil bacteria). Vancomycin very specifically interferes with the cell wall structures of Gram-positive bacteria. This does have rare indications in a maxillofacial clinic.
Susceptible bacteria:
- β-lactam drugs are effective in treating a wide range of infections caused by Gram-positive and Gram-negative bacteria, aerobes as well as anaerobes.
- Vancomycin is effective in treating a range of infections caused by Gram-positive bacteria and has virtually no effect on Gram-negative bacteria (some Gram-positive bacteria are intrinsically resistant to vancomycin, and there is concern over growing numbers of acquired vancomycin resistances of several hard-to-treat bacterial strains). It is reserved for unusual infections in a hospital setting.
- Isoniazid is mainly effective in treating infections caused by mycobacteria tuberculosis – note NOT the atypical mycobacteria often seen in maxillofacial clinics as a cause of neck lumps.
Where these are useful antibacterial agents:
- Owing to the broad-banded properties of the β-lactam antibacterial agents, these are a common first-line choice of treatment for a number of oral and maxillofacial infections, including abscesses, cellulitis, severe alveolar osteitis, acute periodontal disease, necrotising ulcerative gingivitis (although see comments about anaerobicidal drugs), impetigo, erysipelas and actinomycosis.
- Isoniazid, usually in combination with rifampicin (see below), is effective for the treatment of mycobacteria and thus is used for the treatment of tuberculosis.
- Vancomycin is one of the few remaining last-resort antibacterial agents for the treatment of sepsis, severe lower respiratory tract and some bone infections. It also has a limited role topically in the management of antibiotic-induced colitis caused by clostridium difficile.
Agents that interfere with bacterial metabolism
Sulphonamides inhibit the bacterial folic acid synthesis and historically were the first systemically administered broad-spectrum antibacterial agents, initially with spectacular success. They are no longer widely used because of unwanted effects and increasing issues with bacterial resistance (see below). Figure 2 depicts the basic functional unit in sulphonamides in red. Another antibacterial agent which inhibits a different stage of bacterial folic acid synthesis is trimethoprim (see Figure 2).
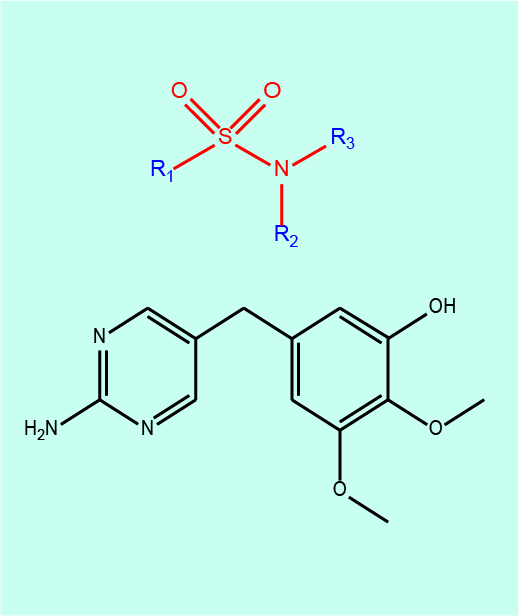
Susceptible bacteria: unless there are resistance issues with sulphonamides (which are common), these include a range of Gram-positive and Gram-negative bacteria, chlamydia, e.coli, klebsiella, salmonella, shigella and enterobacter species.
Where these are useful antibacterial agents: sulphonamides are rarely used as a single agent but trimethoprim, and especially combinations of trimethoprim with a sulphonamide, are still used for the treatment of urinary tract infections.
Agents that interfere with the bacterial protein synthesis
A wide range of different chemical compounds inhibit bacterial protein synthesis by interfering with the bacterial ribosome in various different ways. Figure 3 depicts some examples from this group of different antibacterial agents that are commonly used. These include natural products (for example, chemicals with antibacterial properties which are synthesised by other microbes), semi-synthetic and synthetic compounds.
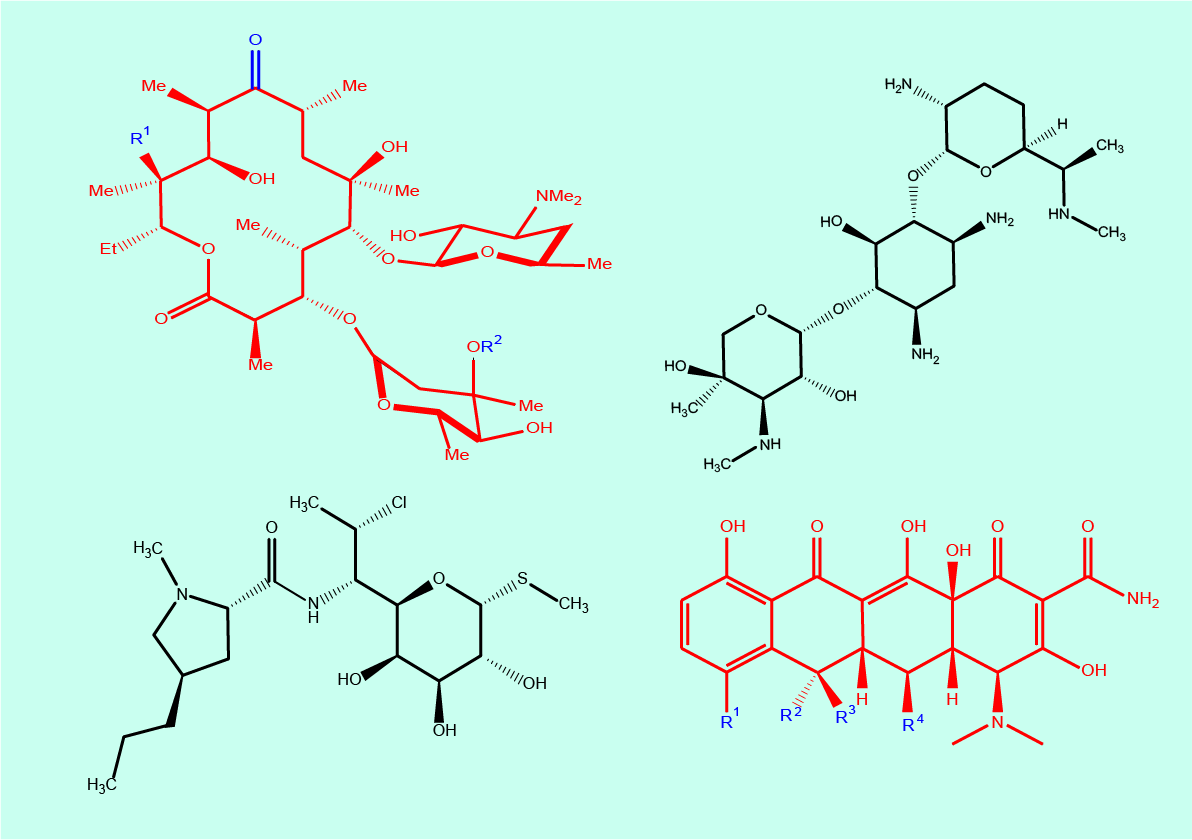
Macrolides (Figure 3 top left) are natural products and include compounds such as clarithromycin and erythromycin. Another semi-synthetic antibacterial agent is clindamycin (Figure 3 bottom left). Gentamicin (and streptomycin) belongs to the chemical class of aminoglycosides (Figure 3 top right). A number of synthetic tetracycline derivatives (Figure 3 bottom right) are another class of antibacterial drugs.
Figure 4 shows yet another antibacterial agent which interferes with bacterial protein synthesis, chloramphenicol.
Susceptible bacteria:
- Macrolides are effective in treating infections mostly by Gram-positive bacteria, including a range of streptococci, pneumococci, staphylococci and enterococci species; macrolides are also effective in the treatment of chlamydia, legionella, mycobacteria and mycoplasma species.
- Clindamycin is effective in treating infections by Gram-positive aerobic bacteria such as most staphylococcus and streptococcus species (many Gram-negative aerobic bacteria (for example, pseudomonas, legionella, haemophilus influenza and some enterobacter species are resistant to clindamycin; see below); clindamycin can also be used for the treatment of infections with some Gram-negative anaerobic bacteria, mostly rod-shaped species.
- Aminoglycosides are effective in treating infections mostly by Gram-negative, aerobic bacteria, including pseudomonas, e. coli, klebsiella and some enterobacter species (some pseudomonas, enterococci and staphylococci species, including staphylococcus aureus, are resistant to gentamicin).
- Tetracyclines started as a group of highly broad-banded antibacterial agents, effective against almost all clinically relevant Gram-positive and Gram-negative bacteria, aerobes and anaerobes. Unfortunately, bacterial resistance (see below) of tetracyclines has become a common feature for a number of staphylococcus, streptococcus, pneumococcus, neisseria and enterobacter species, as well as many anaerobes.
- Chloramphenicol has a broad spectrum of antibacterial activity and is effective in the treatment of infections caused by streptococcus e. coli and staphylococcus (including staphylococcus aureus) species. It is also active against the most common causes of bacterial meningitis, neisseria meningitides, streptococcus pneumoniae and haemophilus influenza.
Where these are useful antibacterial agents:
- Macrolides have a spectrum of activity slightly wider than, but similar to, that of β-lactam antibacterial agents. In a maxillofacial clinic they are a second line drug in the penicillin allergic patient, especially erythromycin is an alternative choice.
- Clindamycin is used for the treatment of dental, respiratory tract and bone infections; it is another alternative choice where people are intolerant of β-lactams. Clindamycin is more widely used in Europe than in the UK where it is rarely used because of its rare but potentially serious unwanted effects: it lost popularity in the UK due to its association with antibiotic-induced colitis.
- Aminoglycosides are mainly used systemically for the initial treatment of serious infections (including respiratory tract infections and sepsis); otherwise it is mainly topical use of aminoglycosides in orthopaedic trauma surgery for the local treatment of bone and soft tissue infections and maxillofacial surgery for osteomyelitis of the jaws. Most aminoglycosides are used in an injection format.
- Tetracyclines remain useful drugs for the treatment of some mixed mouth and throat and some bone and some sinus infections. The latter in particular with regard to infections with meningococci. In addition, tetracyclines get incorporated into living bone tissue. Since the tetracycline molecules can be identified by their fluorescence (a particular stimulated emission of light), they can be used as marker molecules for the identification of healthy and necrotic bone tissues.
- Chloramphenicol easily passes the blood – brain barrier, so has a role in the treatment of brain abscesses and bacterial meningitis in general as a systemic drug. It is not widely used systemically otherwise because of its potentially serious unwanted effects. In a maxillofacial surgery context, the main role of chloramphenicol is as a topical drug, to treat skin infections, to prevent (or treat) infections after skin grafting, and in the form of an ointment for the aftercare of surgical wounds. It is particularly useful as the topical eye ointment comes in a small sterile package for self- application even though that is not what it was designed for. Chloramphenicol is available only as an intravenous and topical preparation.
Agents that interfere with the bacterial DNA
Similar to the antibacterial agents that interfere with bacterial protein synthesis (see above), interference with bacterial DNA synthesis is also a working principle of a wide range of different chemical compounds used as antibacterial agents.
By far the most prominent member of this class of antibacterial agents in maxillofacial surgery is metronidazole (see Figure 5).
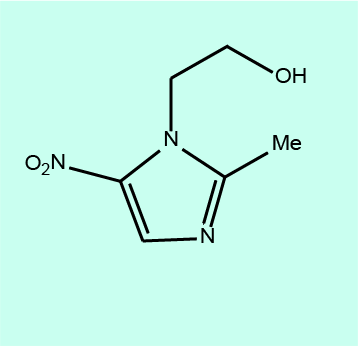
Metronidazole belongs to the class of chemicals called nitroimidazoles; they can be classed as pro-drugs because it is necessary for the NO2 group in the molecule to be chemically reduced (modified) before interference with DNA synthesis can occur.
Rifampicin, a natural product belonging to a group of chemicals called rifamycins (see Figure 6) and the group of fluoro-quinolones (see Figure 7) also interfere with bacterial DNA, though in different ways from the mechanism exploited by metronidazole.
Susceptible bacteria:
- Metronidazole, owing to the requirement of chemical reduction for achieving its antibacterial function, has a wide spectrum of activity in treating infections caused by anaerobes (but has little or no effect on aerobes).
- Rifampicin is effective in treating infections by meningococci, listeria, neisseria and legionella species, as well as haemophilus influenzae.
- F-quinolones are mainly effective in treating infections by Gram-positive bacteria.
Where these are useful antibacterial agents:
- Metronidazole is a widely used, first-line drug in the treatment of many dental, oral and maxillofacial infections, often in combination with β-lactam antibacterial agents (see above) in the treatment of severe infections as they have a synergistic (additive) effect. In addition it is used in the prevention of bacterial infections: it is widely used in a single-shot mode prophylactically in oral and head and neck surgery.
- Rifampicin is not a first-line, widely used antibacterial agent but has several important uses in treating maxillofacial fractures at risk of infection (rifampicin enters the spinal liquid and thus the brain), it usually is part of a combination therapy for treating tuberculosis, it is used for the treatment of some bone and (prosthetic) joint infections, and is sometimes used in the treatment regimen for MRSA infections.
- F-quinolones are not widely used because of their unwanted effects but can be useful in the treatment of chest and urinary tract infections as well as some bone infections such as osteomyelitis (similar to tetracyclines (see above), quinolones are incorporated in bone tissue). If used, they are usually used in combination with other antibacterial agents because many pathogens are resistant to quinolone drugs, or can rapidly acquire resistance even over a short course of treatment.
Antifungal agents
The two most commonly used antifungal agents in maxillofacial surgery are fluconazole and miconazole (see Figure 8 for molecular structures). Both agents hinder the ergosterol synthesis in the fungal cell membrane.
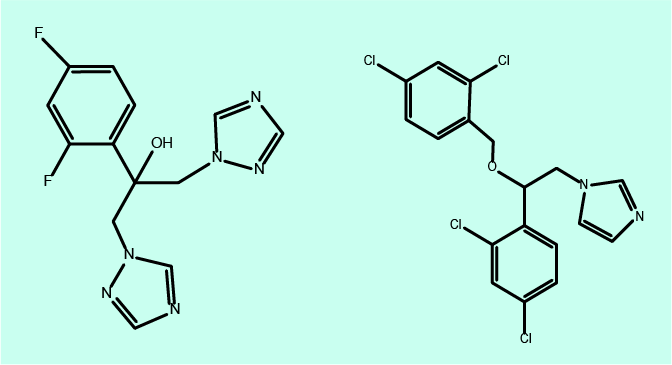
Fluconazole is administered systemically (orally or intravenously), whereas miconazole is used topically (cream on skin, gel in mouth). The majority of maxillofacial fungal infections are caused by candida species, most of which are susceptible to treatment with fluconazole. Where fluconazole fails, fortunately in most cases there is no fungal resistance to miconazole. However, there are some (rare) hard-to-treat fungal infections by candida species which are resistant to first-line antifungal agents.
Antiviral agents
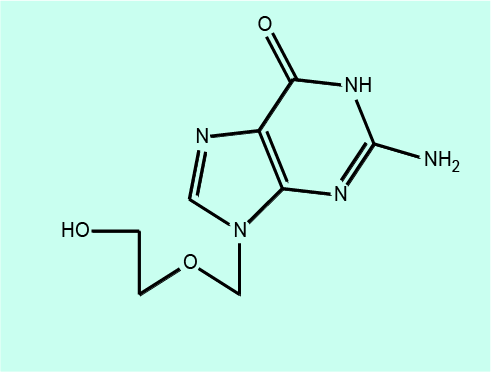
The one antiviral agent that is likely to be prescribed in a maxillofacial clinic is aciclovir, the most commonly used antiviral drug to treat herpes infections (herpes simplex infections and shingles). Aciclovir works by preventing viral DNA replication.
Resistance to antibiotic agents
The developing resistance issues with antibiotic agents are most concerning and most wide-spread as far as antibacterial agents are concerned (although similar issues and mechanisms exist for antifungal and antiviral drugs). Accordingly, most of our discussion will be addressing bacterial resistance.
Resistance to antibacterial agents
With the increased use and overuse of antibacterial agents there has inevitably been the development of bacterial strains, which have resistance to the most commonly used antibacterial agents. Resistance arises when selective pressure is placed upon bacterial colonies by antibacterial agents: most of the bacteria present are sensitive to the antibacterial used; however some remaining (few) bacteria are not eliminated by the antibacterial agent and are then described as resistant. These surviving bacteria can continue to multiply while being unaffected by the continued use of the applied antibacterial agent, without competition from other bacteria. In this way these surviving bacteria are said to have been selected for their desirable resistance traits.
As antibacterial agents are being used increasingly, bacterial strains resistant to multiple antibacterial agents, so called ‘superbugs’ have evolved (much attention has been paid in the media to MRSA in the context of hospital-acquired infections, or more recently to the worrying trend of increasing numbers of mycobacterial infections that cannot be treated by any existing antibacterial agents).
Some bacteria have acquired resistance naturally as they produce antibacterial agents to which their competitor bacteria are susceptible. Many such natural antibacterial agents have been in existence for millions of years (for example, produced by the myriads of useful soil bacteria), it thus makes sense that bacterial resistance mechanisms would have also been necessary to be in existence for a similar length of time.
Figure 10 depicts a cartoon version of a monstrous ‘virtual’ bacterium. The figure symbolises a super-super-bug that has acquired every known trick to the bacterial world to counteract man-made antibacterial agents (to the best of our knowledge, no such super-super-bug exists (yet), but there are several known bacterial strands which use more than one of these mechanisms to counteract the effects of antibacterial agents).
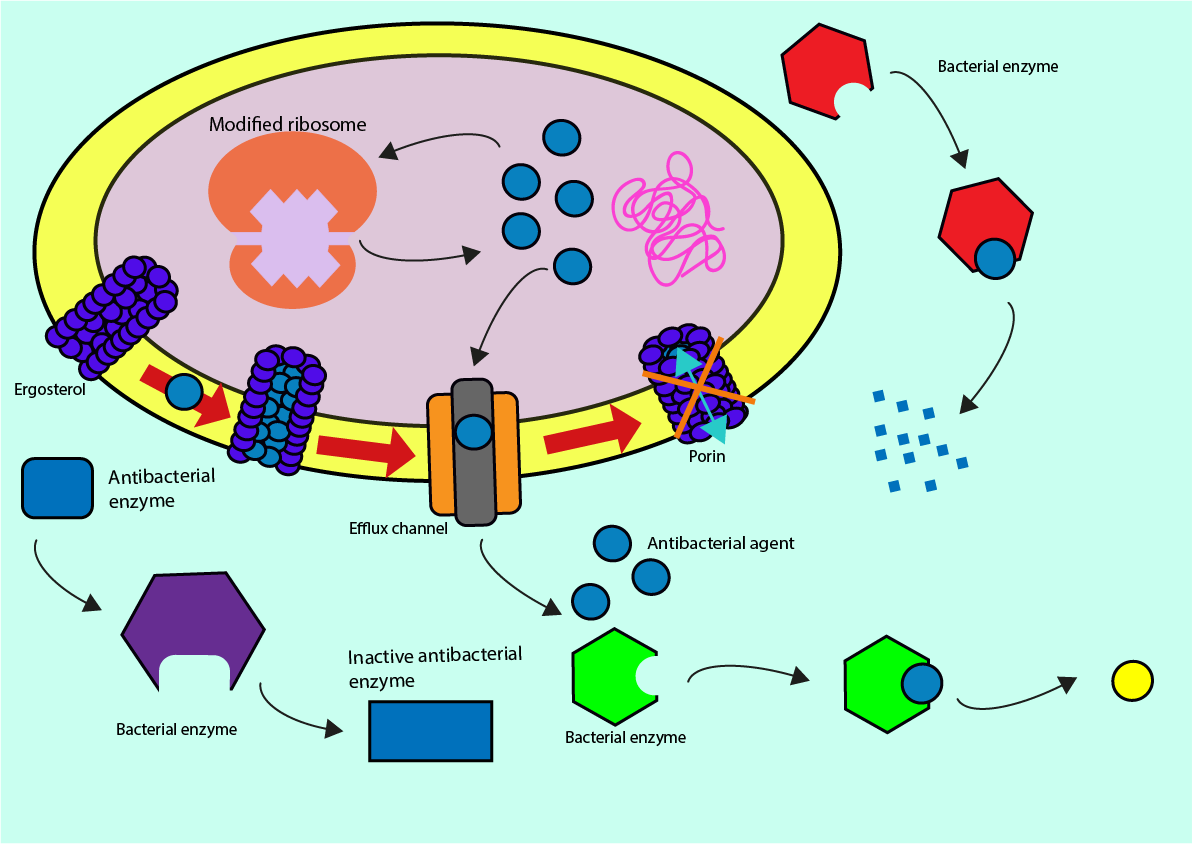
There are four main mechanisms of action which bacteria use in order to resist antibacterial agents:
- altering antibacterial enzymes and antibacterial molecules (reduces complementarity to bacterial molecules and structures)
- breaking down of antibacterial molecules (before they are able to perform their function)
- efflux pumps (swift removal of antibacterial agent from the cell once it managed to get into the cell) and prevention of pore formation (removal or blockage of pathways into the cell for antibacterial agent)
- ribosomal modifications (prevention of antibacterial interference with protein synthesis).
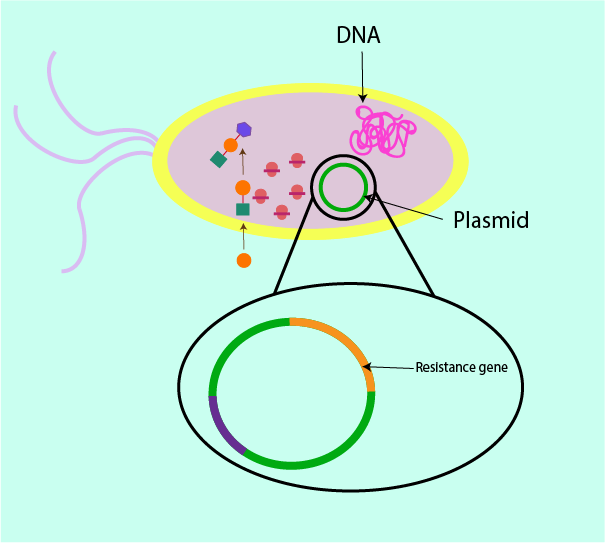
The resistance issue is made worse by the ability of bacteria to pass on their resistance traits and in this way increase the numbers of resistant bacteria on an exponential scale. Resistance genes can be contained on chromosomal (nuclear) or extrachromosomal DNA (plasmid). Figure 11 symbolises the way in which the plasmid can carry resistance genes. Within a bacterial cell, DNA components are also stored on plasmids (circular DNA loops) outside of the nucleus in the cell cytoplasm (extrachromosomal DNA). This plasmid DNA is replicated independently without regulation from the rest of the cell.
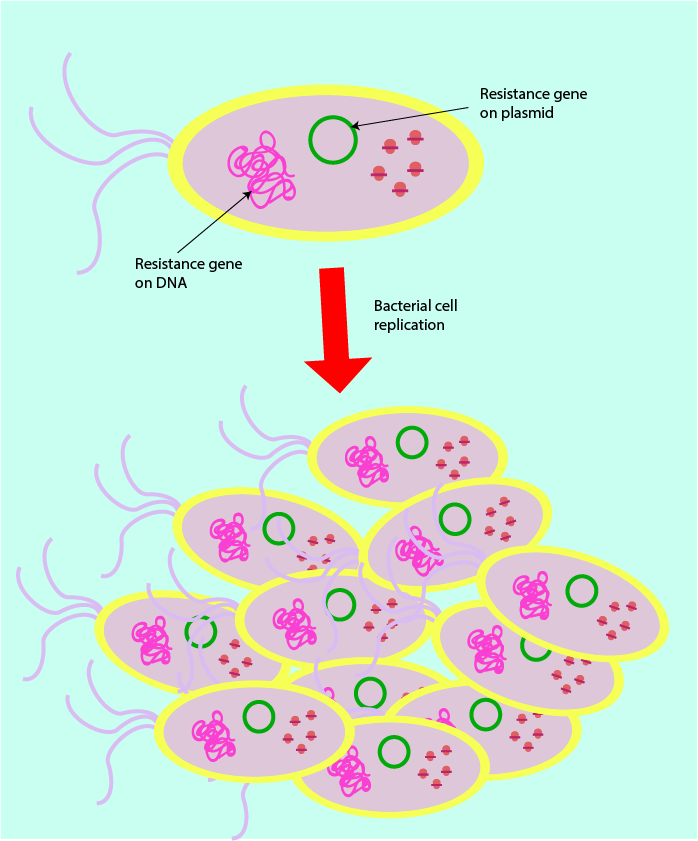
Resistance genes usually arise due to chance mutations of the DNA which provide bacteria with a selective advantage (established when brought into contact with antibacterial agents). The different ways in which resistance genes are passed on are sketched in Figure 12 and Figure 13.
Vertical transmission: resistance is passed on from one generation to the next during bacterial cell replication and DNA replication of the resistance-containing chromosome.
Horizontal conjugation: bacteria form connections (pili) with non-resistant bacteria through which to pass on resistance genes contained on plasmids (extra-chromosomal DNA).
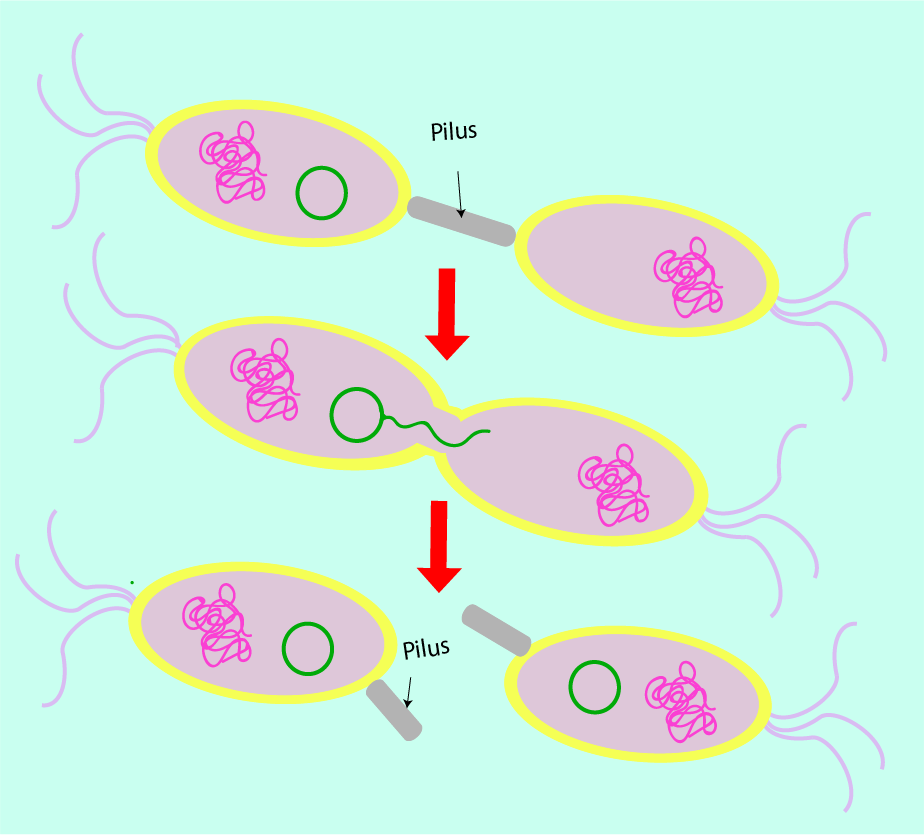
Alteration of antibacterial enzymes and antibacterial molecules (see Figure 10): the bacterial cell may produce molecules to modify and inhibit the antibacterial enzyme so it can no longer carry out its function. The bacterial cell may also apply modifications to its own enzymes (as typically targeted by antibacterials) such as those important in folic acid synthesis or other bacterial biosynthetic pathways. These modifications impact on the shape of the active site and so prevent the binding of inhibitor (antibacterial) molecules. Bacteria may also modify incoming antibacterial molecules such that they cannot bind to the recognition sites of the bacterial cell surface membrane, and so prevent antibacterials from entering the cell altogether.
Break down of antibacterial molecules (see Figure 10): a mutation may result in DNA coding for enzymes which are able to break down antibacterial molecules, preventing them from entering the cell intact and binding with their target, hence causing the antibacterial agent to be ineffective.
Efflux pumps and prevention of pore formation (see Figure 10): some antibacterial agents are able to attach to molecules within the plasma membrane of bacteria and form porins (channels) which make the bacterial cell permeable, leading to eventual bacterial cell death (also a common working principle of antifungal agents). Bacterial cells can be resistant to this effect by making changes to porins and/or introducing efflux pumps into their membranes. The efflux pumps are able to remove antibacterial agent from the cell swiftly once it has been successful in getting into the cell - before the antibacterial is able to perform its function. Efflux is a mechanism commonly employed by most bacteria to reduce antibacterial agent concentration in cells.
Ribosomal modifications (see Figure 10): there are many sites on the ribosome to which antibacterial agents can potentially bind and through which they can interfere with protein synthesis. Advantageous resistance mutations within the DNA, which codes for the proteins which make up the ribosome structure, allows the ribosomal structure to change so as to prevent antibacterial agents from binding. Some bacteria also synthesise proteins, which pass through the binding site of the ribosome and clear out any antibacterial agents, which may have managed to bind successfully to the ribosome (sometimes such proteins are referred to as ‘bottle brushes’).
Some comments about the spread of bacterial resistance (see Figure 11): within the plasmid there may be genes of antibacterial resistance, arising either from a genetic mutation or due to adaptation against naturally occurring antibacterial agents. These are sometimes present in some bacterial cells as a line of defense against competitor bacteria. These resistance genes code for particular proteins. These proteins are then complicit in rendering antibacterials useless through destruction or inhibition through all of the various mechanisms described above.
Of particular concern with regard to antibacterial resistance is the ability of bacterial cells to replicate the plasmid containing the resistance gene and pass it on through conjugation (direct contact) to other bacterial cells, allowing more and more bacterial cells to inherit resistance to certain antibacterial agents. This is a particularly efficient way of spreading resistance amongst bacterial colonies (in addition to the ‘normal’ way of handing genetic information over from generation to generation). The horizontal plasmid-dependent mechanism is mainly responsible for the rapid development and establishment of multiple resistance genes in one bacterial strand, allowing for bacterial strands to develop which are resistant to many different types of antibacterial agents.
Resistance issues have been reported for all classes of antibacterial agents but multiple such issues are becoming more common for many of the first-line, widely used antibacterial agents, including
- β-lactams
- chloramphenicol
- macrolides
- rifamycins
- quinolones
- tetracylines
- isoniazid.